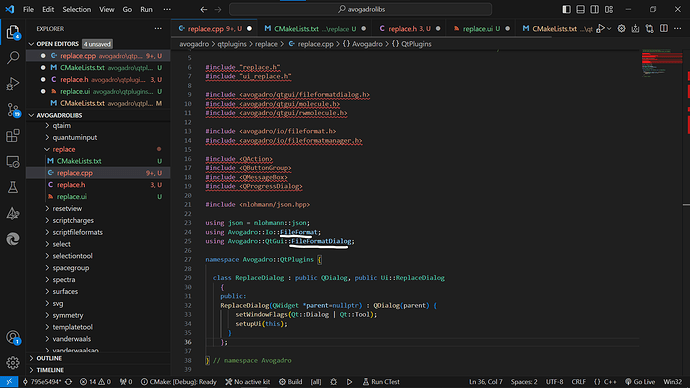
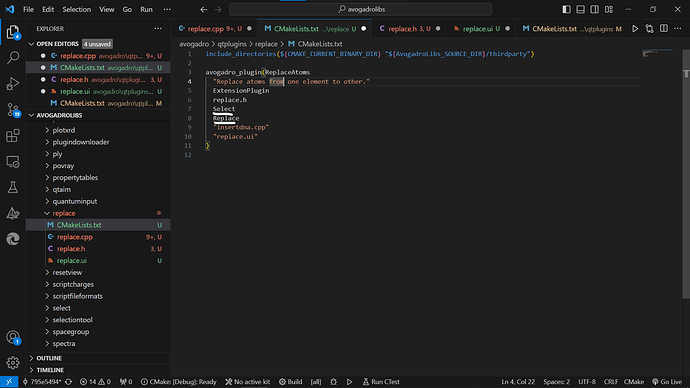
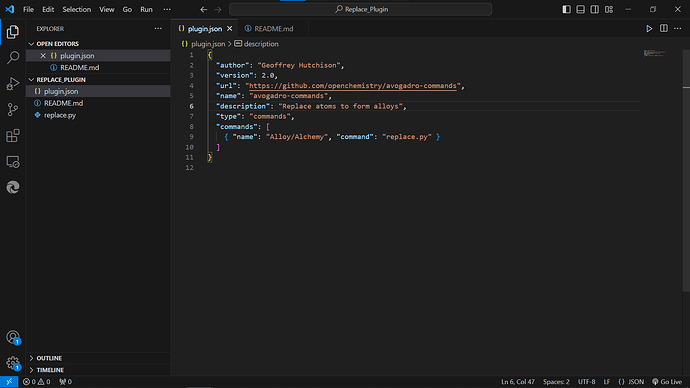
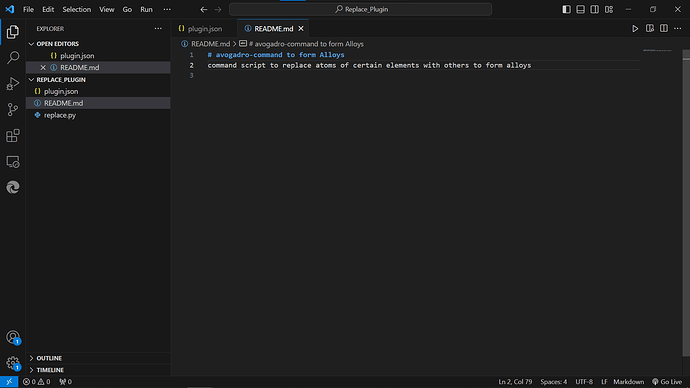
Can you please have a look if its apt
def getOptions():
userOptions = {}
userOptions['Find'] = {}
userOptions['Find']['label'] = 'Find Element'
userOptions['Find']['type'] = 'stringList'
userOptions['Find']['values'] = list(element_dict.keys)
userOptions['Find']['default'] = "Carbon"
userOptions['Percent'] = {}
userOptions['Percent']['label'] = 'Percentage to be replaced'
userOptions['Percent']['type'] = 'float'
userOptions['Percent']['default'] = 100
userOptions['Replace'] = {}
userOptions['Replace']['label'] = 'Replace with'
userOptions['Replace']['type'] = 'stringList'
userOptions['Replace']['values'] = list(element_dict.keys)
userOptions['Replace']['default'] = "Carbon"
opts = {'userOptions': userOptions}
return opts
def replace_element(opts, mol):
original= element_dict.get(opts['Find'])
percent = float(opts['Percent'])
replace= element_dict.get(opts['Replace'])
atomic_numbers = mol['atoms']['elements']['number']
occurences = atomic_numbers.count(original)
number_of_atoms = int((percent*occurences)/100)
indices = []
for i, num in enumerate(atomic_numbers):
if num == original:
indices.append(i)
selected_indices = random.sample(indices, number_of_atoms)
for index in selected_indices:
atomic_numbers[index] = replace
return mol
Output for 2 different instances for input [1, 1, 8, 1, 9, 1] 25% of hydrogen atoms to be replaced randomly with carbon
{"cjson": {"chemicalJson": 1, "atoms": {"coords": {"3d": [0.617532, -0.027246, 0.481009, 0.156663, 0.031752, -0.362419, -0.774185, -0.004516, -0.11859]}, "elements": {"number": [1, 6, 8, 1, 9, 6]}}, "bonds": {"connections": {"index": [1, 2, 1, 0]}, "order": [1, 1]}}}
{"cjson": {"chemicalJson": 1, "atoms": {"coords": {"3d": [0.617532, -0.027246, 0.481009, 0.156663, 0.031752, -0.362419, -0.774185, -0.004516, -0.11859]}, "elements": {"number": [1, 1, 8, 6, 9, 6]}}, "bonds": {"connections": {"index": [1, 2, 1, 0]}, "order": [1, 1]}}}
Also , shouldn’t this line be elif args['run_command']: