Hello Sean,
sorry for the delay, I hope this time the message reaches you promptly.
I wasn’t yet aware about moltemplate - I assume it is the project you refer to. However, with some of the illustrations from this repository in hand, and your description, the reasoning is evident.
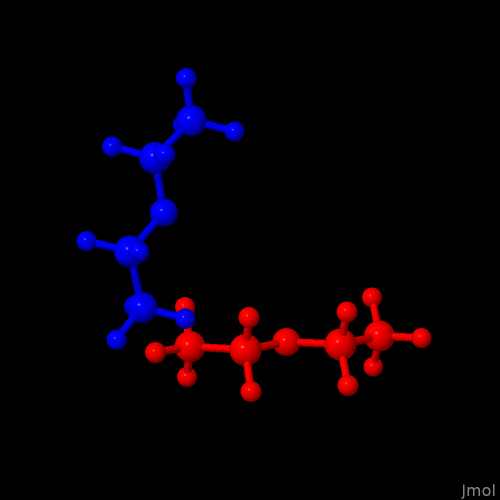
For now, I identified orient-molecule as a handy implementation which can translate and rotate/reorient the molecules. Based on a test with an openbabel generated model of diethyl ether, the Python based utility (padded by numpy) seems to work e.g., by
python3 ./orient.py -rz 90 ./diethylether.xyz > rz90.xyz
The simultaneous display of both the original as well as the modified model file by Jmol yields this:
The attached markdown log describes the individual steps. Right now, I do not recall if openbabel’s documentation includes a diagram about the coordinate system’s handiness. Do you know if there is a convention?
log.md.txt (1.4 KB)