Summer’s over, so Google Summer of Code comes to an end. As a participant, I feel I’ve learned many useful skills during these months (also had lots of fun!). Let’s review the improvements I’ve made to Avogadro, what I’ve been working on this summer. I’m glad to present…
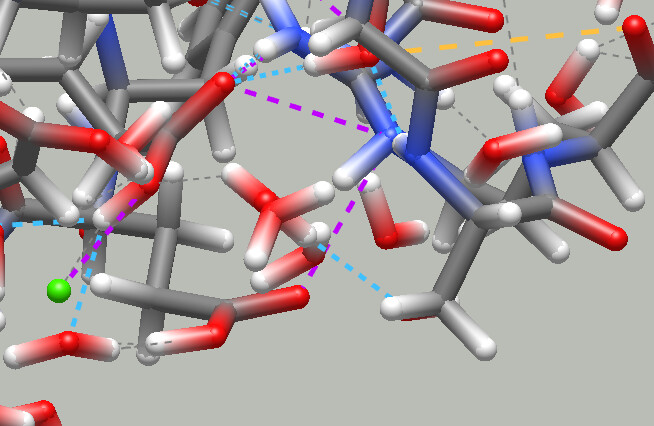
Non-covalent interaction rendering
Avogadro 1.96 introduces “close contacts”, which show dashed lines between atoms that are closer than some threshold. In Avogadro 1.97 this is expanded to include salt bridges, “bad” contacts (repulsive forces between same-sign charges), as well as hydrogen, chalcogen and halogen bonds, which take angles between electron pairs into account for better prediction.
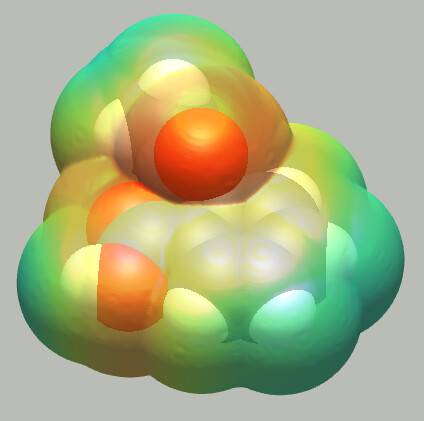
Molecular surfaces
With Avogadro 1.97 finally comes the ability to compute surfaces for a molecule. Van der Waals, Solvent Accessible and Solvent Excluded surfaces are supported, at various resolution and smoothing levels. Additionally, surfaces can be colored by electrostatic potential, also allowing to choose the model to use.
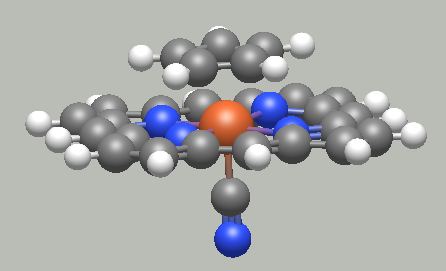
Template tool
@ghutchis and I worked together to add a tool that allows one to easily create coordination complexes, or add fuctional groups to an organic molecule. Polydentate and haptic ligands are properly aligned around the selected geometry. This tool will be availbale in 1.98.
Real-time ambient occlusion
This wasn’t actually part of my original schedule, but I had spare time so added it as a bonus feature. A new shader pipeline is now in place to add real-time ambient occlusion and edge detection, so complex molecules are better visualized. A dialog offers configuration options over these shaders. This will appear in 1.98.
Performance changes
While not offering any extra functionality, these are important to keep working with very large molecules feasible, especially on slower machines. During this summer I optimized PDB import to be tens of times faster, as well as some nice performance figures for other operations (e.g. adding hydrogens, enabling cartoons…). Most of this work is already in 1.96.
So, that’s it! I hope the Avogadro community will find some nice uses for these new features. I’ve mostly focused on uses for biochemistry (as I’ve used Avogadro to better understand it myself, reminder that I’m a med student!), but I hope it benefits everyone in some way. Thanks for the opportunity and for this wonderful piece of software!