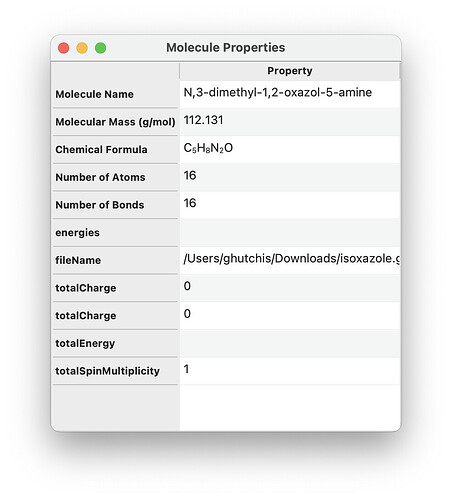
Looks good!
Agreed.
I also don’t think this is the right place for the list of energies, so I would leave them out of here. There’ll be a conformers window or something for it to go in where it is more clear what they correspond to.
Making as much as possible translatable is important in my view so +1.
Please please please take the time to make this dependent on a setting in the user’s config. It doesn’t necessarily need to be possible yet to actually change that setting in the UI, but I’d beg you to set up a config option now in advance, and refer to that for the molecular properties dialog.
Ideally all energies across the whole program should then be shown in the user’s preferred units. It can be overruled in individual cases if there is a very good reason to use different units for them.
For this molecular properties dialog in particular I would add a third column for units, and in some cases I would let this be a drop-down to let the user temporarily switch the units for a given field. Then everything is super convenient – the user can set their preferred global default units, yet also get values in other units when necessary without having to change the global setting or get out a calculator.
To cover different disciplines, the options should be at minimum:
- kJ/mol
- kcal/mol
- hartree (not capitalized, like kelvin, ohm etc.)
- eV
- cm-1
Only for computational chemists! School and undergrad students have no idea what a hartree is.
Besides, reaction/transition state energies etc. are always in kJ or kcal, so in my personal experience it’s rare that a single point energy in hartrees is useful as it always needs converting.
Yes, but most people aren’t computational chemists  Even leaving aside the fact that kcal is a stupid unit, it’s use is mostly restricted to chemistry and even within that is most prevalent in comp chem. Avogadro should be broader than that. And I know in the US everyone still uses kcal/mol for e.g. bond enthalpies, but I can tell you that in the UK all chemical education at all levels speaks in kJ/mol. I only had to deal with kcal when I went abroad, and British comp chemists end up switching during their PhD lol.
Even leaving aside the fact that kcal is a stupid unit, it’s use is mostly restricted to chemistry and even within that is most prevalent in comp chem. Avogadro should be broader than that. And I know in the US everyone still uses kcal/mol for e.g. bond enthalpies, but I can tell you that in the UK all chemical education at all levels speaks in kJ/mol. I only had to deal with kcal when I went abroad, and British comp chemists end up switching during their PhD lol.
You both are computational chemists so no doubt have different experiences and needs, but that’s my point – different users need different units 
IMO kJ should be default, because SI and IUPAC, but as long as it can (at some point) be changed I don’t mind hugely.
This information is not currently stored within the CJSON anywhere, so I imagine it would be quite complex and quite a change to store it for each value. It is easier to store in hartree (like now) and convert on-the-fly for display. And I think users would rather get the result in the units they like rather than the units that the orca/gaussian/turbomole/xtb/molden/gamess/quantum espresso authors like, especially people who work with more than one program!