Hello,
I am trying to visualize the MOs from my ORCA output. While Avogadro 1.97 can open my file through cclib, it doesn’t display the MOs. To address this, I used the orca_2mkl utility to convert my output to a Molden file.
However, when I attempted to open the Molden file in Avogadro, I encountered an issue. Subsequently, I tried using the ccget --list command with the ORCA 5.0 output and had no problems. However, when I used this command with my converted file, cclib 1.8 failed to work.
You previously mentioned in another thread that the Avogadro2 worked well to open converted files. So, I’m wondering if there might be an issue with my specific molecule/Avogadro 2, or if cclib 1.8 has a bug.
Best regards
3_21G_molden.out (166.8 KB)
3_21G.out (513.8 KB)
First off, Orca does not always include MO information in the output. To do that, you need something like this in the input files:
%output
print[p_mos] true
print[p_basis] 5
end
If the calculation was run without those directives, there’s nothing Avogadro or cclib can do.
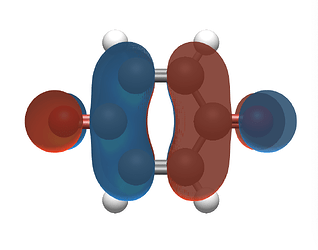
I can confirm that Avogadro 1.98 reads your Orca output with no problem:
As far as the Molden… I think you’ll have better luck if you use .molden as the extension. (And I’m pushing a code change to check for Molden files with .out for 1.98)
Hope that helps - are you able to use an AppImage build?
Hello,
Thank you for your reply and time.
In the command line of the file I provided, I included “PrintMos” and “PrintBasis,” and I believed that it would be sufficient. However, even after using your suggested keywords, when I load the file (generated with your added keywords), Avogadro2 displays the molecule but not the orbitals toolbar. Nevertheless, “ccget --list 3_21G.out” now works perfectly fine.
Does this mean that Avogadro 2 cannot display orbitals unless the file is transformed into a Molden file, or did I make an error in my approach?
I do not use an AppImage build. I git clone the depository and install everything manuall
3_21G.out (513.8 KB)
I also noticed that when I opened the corresponding Molden file (I changed the .out by .molden), the orbitals were not displayed. I feel quite confused because you were able to plot them, whereas I don’t see any windows.
Try the 1.98 release. Note that the orbitals won’t show up automatically- you need to use Create Surfaces
Hello,
I have installed the latest version of Avogadro, but I still cannot see the orbitals. For some reason, I am unable to choose the reader as before (cclib, OpenBabel, or Avogadro). I still have cclib, and it is in the same folder as my previous version of Avogadro (1.97). So, I do not understand why the upgrade caused this issue.
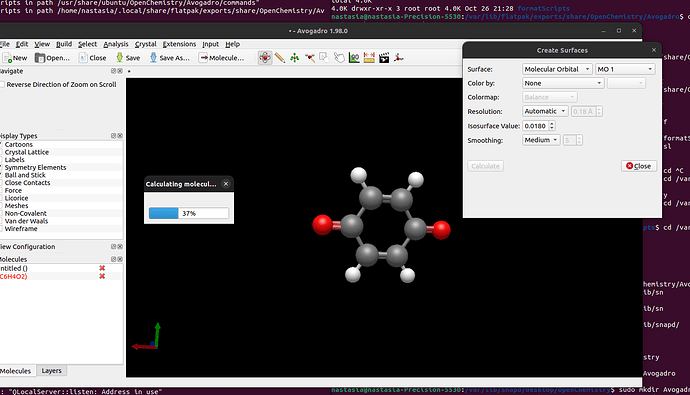
I am able to open my molecule, and I clicked on “Create Surfaces” → “MO.” However, I do not understand why it is computing them, whereas they should be in the output. Plus, no orbitals are shown at the end of the Avogadro calculations.
By popular request, that dialog was made optional. If you really want to choose a reader, you can hold down the Shift or Alt / Option key when you open the file. It will choose intelligently - internal code, then scripts like cclib then Open Babel.
I don’t know what you mean. The output file includes the MO coefficients and basis set. Avogadro still needs to calculate the orbital functions (from the atomic positions, MO coefficients and basis set) and the relevant isosurface to mesh. Gaussian, for example would generate a “cube” file, which contains the orbital values … and thus the isosurface is easy to generate from that.
In Avogadro v1.2, we had a window that would come up and start to pre-compute 5 occupied and 5 unoccupied orbitals. That feature hasn’t been ported, so you still need to use the Create Surfaces command.
As far as why it’s not appearing? It looks like you have turned off the “Meshes” display type on the left of your screenshot.