Hello,
I would like to generate a file for docking (with SwissDock). I understand that the MOL2 file will have to have “all hydrogens and 3D coordinates” . Can I prepare such a file with Avogadro and how?
I tried with Build>Hydrogens>adjust(or add) Hydrogens but nothing happened.
What would the right format for MOL2? “Molprot input format”?
Thank you
How do you generate the structures in first place? Do you use Avogadro as a sketcher? If I do (version 1.97, an AppImage for Linux), whenever I tap (left mouse button) to an other place, the carbon atom previously drawn gets all its four hydrogens I anticipate:
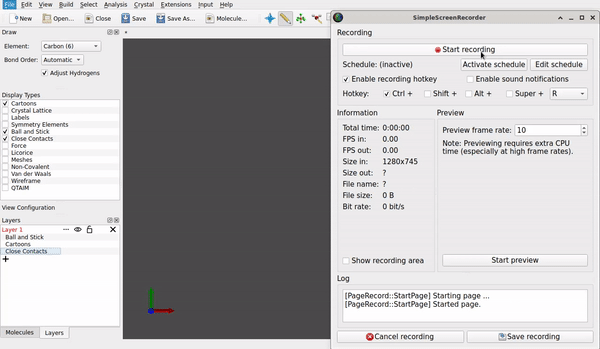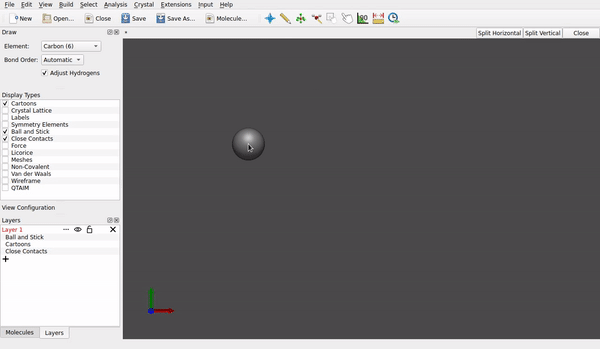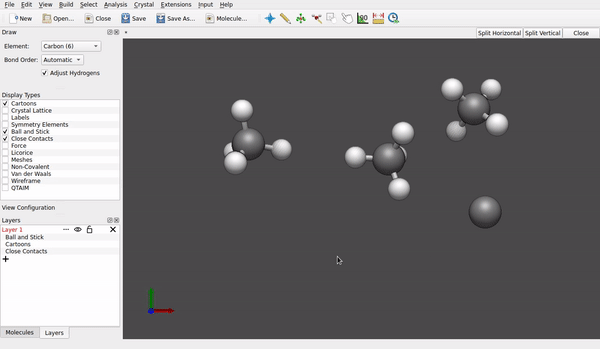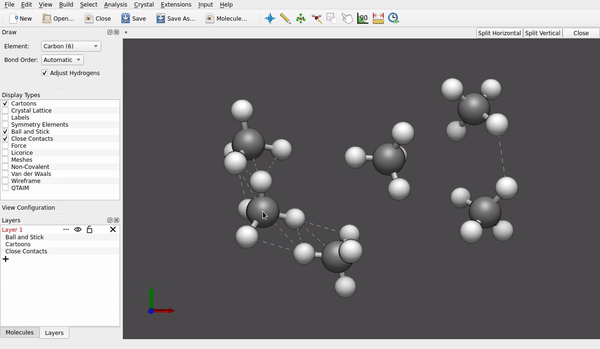
And if I don’t, and just proceed with File → Export → Molecule → Sybyl .mol2 format with a file name like test.mol2, the structures normally include both the framework of carbon/nitrogen/oxygen atoms and peripheral hydrogen, too.
Or, do you use Avogadro as a converter, because you already have structures of interest, however in a other file format than .mol2?