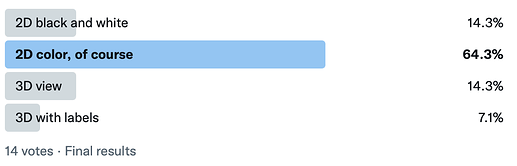Okay, I’ve been working on a “build by fragment” tool, that will also eventually support attaching bidentate and tridentate ligands, π-conjugated ligands like Cp rings, etc.
I started with a list of ligands in a pop-up menu, but it seemed like images would be good.
My thought is that you’d pick a ligand and have a preview on the right side, but I’m curious whether it’s better to have a 2D depiction or a 3D depiction.
Thoughts? Feedback?
- No pictures, list of names is okay
- 2D black-and-white “ChemDraw” style
- 2D color “ChemDraw” style
- 3D Balls-and-Sticks style
- 3D Balls-and-Sticks style with atom labels
- Something else (please comment below)
Since I haven’t gotten many responses on here, I also created a Twitter poll:
https://twitter.com/ghutchis/status/1439995620400537609
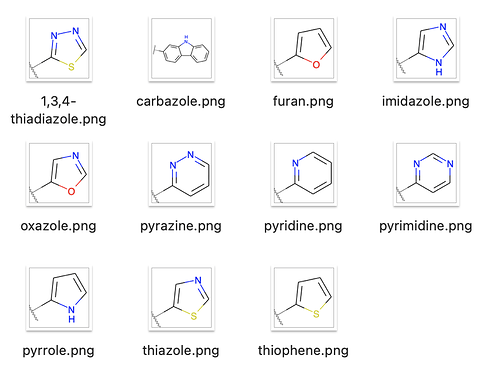
Twitter seems strongly in favor of color 2D diagrams, so I coded up preview images in RDKit today.
I have 113 fragments so far:
- alkanes
- aromatics
- cyclic alkanes
- bicyclic
- heteroaromatics
- boron
- oxygen
- sulfur
- nitrogen
- phosphorus
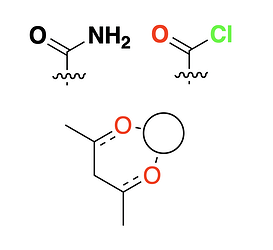
Here are a few 2D heterocycles:
The code generates all the fragments from SMILES, so it should be pretty easy to add / tweak them over time.
I’ll also generate 3D preview images with atom labels, but that needs some separate work (e.g. adding support to tell Avogadro to save an image to a particular filename).