I’m trying to find the conformers for alanine dipeptide and get xyz outputs for each to input into Orca to find energies, etc.
I previously tried on Avogadro 1.2.0 but it was saying that there were no rotatable bonds.
When using conformer search on Avogadro2 (nightly), I think it did find conformers because the molecule itself did rotate in its bonds. I’m not sure where to find these conformers to get the XYZ files. I have seen the command line inputs on other posts but I don’t know where to find this.
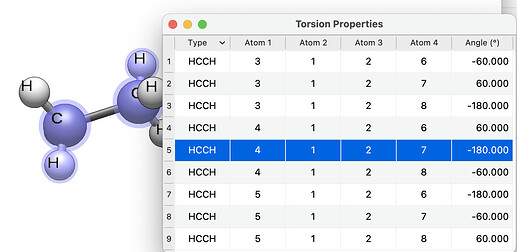
I’m also wondering if there would be a way to find only a few important angles as I do not want to be calculating for a whole bunch of conformers.
Or if there was a way to calculate properties with these conformers in Avogadro.
coordinates I’m using:
C -2.88167 0.36820 -0.10783
C -1.73195 1.25158 0.29351
C 0.75359 1.41582 0.32917
C 0.88829 2.77998 -0.34293
C 1.91566 0.46949 -0.05236
C 4.35891 0.16688 -0.02799
N -0.49752 0.75382 -0.03013
N 3.17785 0.95471 0.21840
O 1.71714 -0.65087 -0.52413
O -1.90588 2.32750 0.85813
H -0.40885 -0.19702 -0.38024
H 3.27485 1.80184 0.76328
H 4.28476 -0.33525 -0.99622
H 4.45501 -0.57874 0.76524
H 5.22614 0.83158 -0.01823
H 0.76513 1.52890 1.41975
H 1.90397 3.18052 -0.27913
H 0.22737 3.51224 0.13195
H 0.61655 2.72711 -1.40271
H -2.53796 -0.54715 -0.59714
H -3.52797 0.91145 -0.80264
H -3.44660 0.09398 0.78698