Hey everyone,
I am a new user of Avogadro so maybe I am missing something evident.
Here is my problem :
For my PhD I want to build a 3D model of the nanoparticle I work with. It is quite a big one around 15 kDa. I can peform the construction to the end but when I save the file I can’t open it again, I have this message instead : “Reading molecular file failed”
I get this message when the “cml” file start to be bigger than 100 Ko.
Do you have any idea ? Is there a file size limit on Avgadro ? Maybe be I can save it in an other format to build it somewhere else ?
Thank you for your help !!
Do you need to start with a .cml file about a structure of 15 kDa? What if you import the structure in a different format (e.g., .pdb for RNA/DNA, if .pdb was the original file format), or/and with a structure with less than 15 kDa to start with? Are there repetitive units like unit cells which 1) one could load, and 2) then multiply by translation in Avogadro? If you construct a nanoparticle with Avogadro, what is the target file format you want to work with after using Avogadro? (I assume you have access to Avogadro 1.95 as provided e.g., by Debian (tracker).)
If the input file is not confidential, you may consider to deposit an accessible copy e.g., on pastebin.com.
Avogadro works with lots of different file formats. It might help if you can upload the file somewhere and we can take a look.
I’m not aware of a limit to CML files, although I admit I haven’t built any 15 kDa nanoparticles. ![]()
If you want a different file format, I might suggest XYZ which should be essentially limitless.
hi Geoff, I ran into this opening .pdb file issue with Avagadro as well (file attached).
4f5s.pdb (1.6 MB)
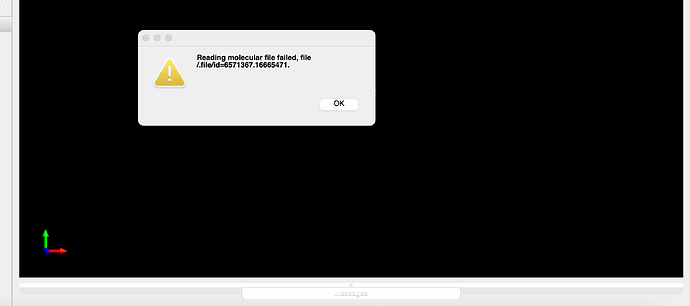
It says “reading multiple molecule files” for a sec and reports error as “reading molecular file failed, file/.file/id=6571367.166665471.” (see attached screen shot). Anything weird?
I tried jmol, the file seems okay, not so sure why Avagadro failed. Thanks a lot!
I can see that you’re using Avogadro v1.2. We can’t support that anymore - it was released over 8 years ago.
I’d suggest grabbing the latest nightly build from https://two.avogadro.cc/ - it works fine (in addition you can directly grab 4f5s from the PDB:
hi Geoff, thanks a lot! I stick to this old Avagodro since it can load Orca computation results including frequency calculation for visualisation purposes.
Is there a new Avogadro version for Macbook that now can support Orca with vibrational mode visualisaiton? The last time (a few months ago), I tried lastest Avogadro, and find it fails to visualise Orca frequency computation results.
Thanks a lot.
I took your advice, it works well! just one question on Avogadro 2 (it shows 1.99.0 though), is there any reason for IR spectra, absorbance conversion were taken out but kept only transmittance in the new version? thanks.
At the moment, I’m the main developer, so progress on the spectra support is slow - I had a busy semester.
If you have other questions about 1.99 (i.e., a beta for 2.0) please post them in a new thread, thanks.