Hi everybody!
I recently got and learned how to use Avogadro, and while I’m overall pretty impressed by its capabilities and intuitiveness, I’ve run into some things that I’m not sure how to do and don’t seem to be covered in the manual.
I’m currently trying to build a thiol self assembled monolayer (SAM) system on gold, and while I can easily create the Au (111) surface, the proper unit cell for the thiol surfactants, and orient them in a way that’s reasonable, I’m not certain how to actually bind the sulfur headgroups to the gold surface.
Additionally, the Au (111) slab is a triangular surface, and while I can see that this makes sense from a crystallographic point of view, I’m not sure how to make that work when I need to impose boundary conditions in the xyz directions for the MD simulations I want to run.
Any help would be greatly appreciated!
I guess you can just build a bond between your thiol group and Au surface. Just don’t forget to unselect “Adjust Hydrogens” before making a bond. If you use version 1.2 then you can optimize the resulting structure fixing the Au surface (select Au atoms, click Extensions – Molecular mechanics – Fix selected atoms).
I did it for the mock Au surface:
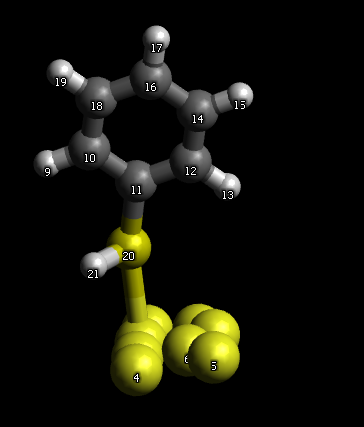
As for the boundary conditions – I am not a pro in MD programs, but I guess that only cubic PBC is usually available. A triclinic cell can be built in GROMACS, but I am not sure it fits your goals. In any case I guess you will have to input cell sizes and angles in your MD program manually.
My suggestion if you’re going to do this on a regular basis, is to use either ASE or pymatgen.
ASE
ASE example: https://wiki.fysik.dtu.dk/ase/ase/build/surface.html
From John Kitchen CMU: http://kitchingroup.cheme.cmu.edu/dft-book/dft.html
Zimmerman group Michigan: https://sites.lsa.umich.edu/zimmerman-lab/tutorial/surface-growing-string-method/surface-setup/
Pymatgen
Matgenb: http://matgenb.materialsvirtuallab.org/2018/07/24/Adsorption-on-solid-surfaces.html
In short, you can create your thiol in Avogadro, save the coordinates for running ASE or Pymatgen. The examples illustrate taking a molecule as a list of atoms in Python.
My hope is that Avo2 will have some tools to interface with ASE and Pymatgen for doing these, but it’s obviously not here yet.
