Dear Avogadro developers and users,
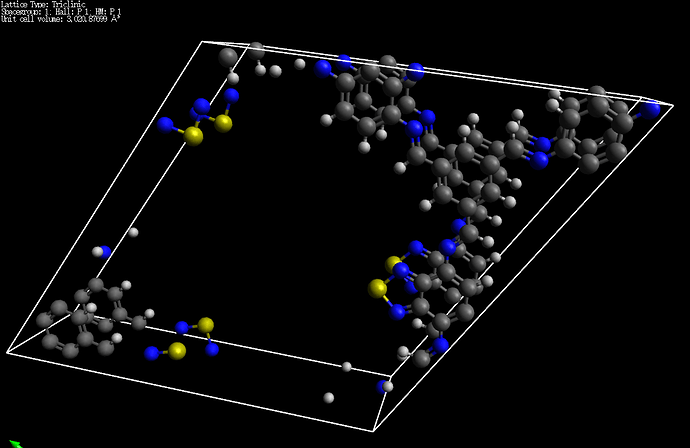
I’m working on building hypothetical COFs. The structure in [112] lattice seems well with cif. or pdb. input. However, the bond type has been changed when I tried to replicate it using supercell builder (for example the double bonds became single bonds). So I was wondering how to solve this problem? Thanks in advance.
I’d be very curious to see a specific example in which the double bond became a single bond.
We can’t maintain Avogadro 1.x. Several people have requested a better experience in the supercell builder for Avogadro 2, including not necessarily “filling the cell” as you depict here.
Besides filling the cell, IIRC the code just copies bonds from the original set of atoms.
If you can give a specific file, it would certainly be helpful when I get to that code with Avo2.
Sure, the input file I used is as follows. Thank you for your quick reply. (sorry for the mess, cuz cif format is not authorized)
data_cif
_audit_creation_method ‘generated by GULP’
_symmetry_space_group_name_H-M ‘P 1’
_symmetry_Int_Tables_number 1
_symmetry_cell_setting triclinic
_cell_length_a 23.9129
_cell_length_b 20.7511
_cell_length_c 6.9720
_cell_angle_alpha 90.0356
_cell_angle_beta 90.0013
_cell_angle_gamma 63.8494
loop_
_atom_site_label
_atom_site_fract_x
_atom_site_fract_y
_atom_site_fract_z
_atom_site_occupancy
C1 0.99654 0.99471 0.24994 1.0000
C1 0.95337 0.96567 0.25005 1.0000
C1 0.97687 0.89081 0.25020 1.0000
C1 0.93654 0.85788 0.25011 1.0000
C1 0.95912 0.78319 0.25006 1.0000
C1 0.87199 0.90091 0.25000 1.0000
C1 0.84684 0.97660 0.24995 1.0000
C1 0.78225 0.02155 0.24988 1.0000
C1 0.88787 0.00830 0.24995 1.0000
H1 0.96683 0.05015 0.24912 1.0000
H1 0.02693 0.85825 0.25028 1.0000
H1 0.92554 0.76181 0.24998 1.0000
H1 0.84117 0.87512 0.24996 1.0000
H1 0.76389 0.98403 0.24974 1.0000
H1 0.86860 0.06644 0.24989 1.0000
C1 0.21911 0.15612 0.24974 1.0000
C1 0.25177 0.19811 0.24987 1.0000
C1 0.22054 0.27319 0.24996 1.0000
C1 0.25352 0.32142 0.24999 1.0000
C1 0.21877 0.40680 0.24999 1.0000
C1 0.32388 0.28036 0.24999 1.0000
C1 0.35895 0.19864 0.24994 1.0000
C1 0.43238 0.15092 0.24992 1.0000
C1 0.31674 0.16238 0.24988 1.0000
H1 0.24625 0.09888 0.24930 1.0000
H1 0.17006 0.29969 0.24996 1.0000
H1 0.23528 0.36344 0.24999 1.0000
H1 0.35084 0.31076 0.25000 1.0000
H1 0.39587 0.17939 0.24993 1.0000
H1 0.33952 0.10413 0.24986 1.0000
C1 0.99626 0.00060 0.74987 1.0000
C1 0.95440 0.96987 0.75001 1.0000
C1 0.97944 0.89467 0.75017 1.0000
C1 0.94031 0.86015 0.75009 1.0000
C1 0.96391 0.78523 0.75005 1.0000
C1 0.87555 0.90194 0.74999 1.0000
C1 0.84915 0.97770 0.74994 1.0000
C1 0.78450 0.02149 0.74988 1.0000
C1 0.88890 0.01098 0.74994 1.0000
H1 0.96610 0.05613 0.74912 1.0000
H1 0.02964 0.86315 0.75024 1.0000
H1 0.93066 0.76342 0.74997 1.0000
H1 0.84550 0.87517 0.74995 1.0000
H1 0.76685 0.98320 0.74974 1.0000
H1 0.86868 0.06915 0.74989 1.0000
C1 0.22130 0.15785 0.74980 1.0000
C1 0.25442 0.19937 0.74989 1.0000
C1 0.22366 0.27452 0.74996 1.0000
C1 0.25714 0.32228 0.74998 1.0000
C1 0.22306 0.40773 0.74999 1.0000
C1 0.32742 0.28060 0.74999 1.0000
C1 0.36198 0.19885 0.74994 1.0000
C1 0.43532 0.15070 0.74992 1.0000
C1 0.31935 0.16311 0.74990 1.0000
H1 0.24806 0.10051 0.74946 1.0000
H1 0.17319 0.30142 0.74996 1.0000
H1 0.23910 0.36432 0.74998 1.0000
H1 0.35476 0.31057 0.75000 1.0000
H1 0.39887 0.17925 0.74994 1.0000
H1 0.34180 0.10484 0.74988 1.0000
N1 0.98487 0.06691 0.24922 1.0000
C1 0.03027 0.09426 0.24954 1.0000
C1 0.00989 0.16877 0.24823 1.0000
C1 0.05286 0.19790 0.24840 1.0000
C1 0.11736 0.15340 0.24982 1.0000
N1 0.15592 0.18610 0.24966 1.0000
C1 0.13693 0.07813 0.25137 1.0000
N1 0.19580 0.02669 0.25349 1.0000
S1 0.19991 0.94321 0.25500 1.0000
N1 0.12135 0.97806 0.25294 1.0000
C1 0.09463 0.05002 0.25116 1.0000
H1 0.96048 0.20441 0.24699 1.0000
H1 0.03564 0.25572 0.24726 1.0000
N1 0.02119 0.73963 0.25006 1.0000
C1 0.05201 0.66660 0.25000 1.0000
C1 0.11747 0.63549 0.25000 1.0000
C1 0.15304 0.56073 0.24998 1.0000
C1 0.12359 0.51489 0.24995 1.0000
N1 0.15599 0.44224 0.24995 1.0000
C1 0.05853 0.54650 0.24992 1.0000
N1 0.02419 0.50995 0.24988 1.0000
S1 0.94705 0.56576 0.24987 1.0000
N1 0.96226 0.63806 0.24993 1.0000
C1 0.02353 0.61950 0.24995 1.0000
H1 0.14120 0.66949 0.25003 1.0000
H1 0.20332 0.53917 0.24999 1.0000
N1 0.73334 0.00186 0.24978 1.0000
C1 0.66788 0.04813 0.24985 1.0000
C1 0.62670 0.01662 0.24963 1.0000
C1 0.56210 0.05912 0.24968 1.0000
C1 0.53702 0.13495 0.24997 1.0000
N1 0.47453 0.17787 0.25004 1.0000
C1 0.57913 0.16517 0.25020 1.0000
N1 0.56321 0.23570 0.25052 1.0000
S1 0.62693 0.25155 0.25076 1.0000
N1 0.67471 0.16226 0.25042 1.0000
C1 0.64235 0.12348 0.25015 1.0000
H1 0.64479 0.95874 0.24939 1.0000
H1 0.53220 0.03231 0.24948 1.0000
N1 0.98449 0.07270 0.74923 1.0000
C1 0.03051 0.09911 0.74955 1.0000
C1 0.01098 0.17344 0.74827 1.0000
C1 0.05461 0.20167 0.74842 1.0000
C1 0.11895 0.15645 0.74983 1.0000
N1 0.15810 0.18842 0.74966 1.0000
C1 0.13776 0.08137 0.75136 1.0000
N1 0.19640 0.02937 0.75344 1.0000
S1 0.19962 0.94630 0.75498 1.0000
N1 0.12099 0.98211 0.75295 1.0000
C1 0.09485 0.05408 0.75117 1.0000
H1 0.96174 0.20967 0.74704 1.0000
H1 0.03800 0.25938 0.74729 1.0000
N1 0.02612 0.74191 0.75006 1.0000
C1 0.05695 0.66871 0.75000 1.0000
C1 0.12248 0.63722 0.75000 1.0000
C1 0.15791 0.56230 0.74998 1.0000
C1 0.12824 0.51666 0.74995 1.0000
N1 0.16031 0.44393 0.74995 1.0000
C1 0.06322 0.54856 0.74992 1.0000
N1 0.02874 0.51218 0.74988 1.0000
S1 0.95168 0.56814 0.74988 1.0000
N1 0.96708 0.64029 0.74993 1.0000
C1 0.02837 0.62163 0.74995 1.0000
H1 0.14642 0.67101 0.75003 1.0000
H1 0.20821 0.54046 0.74999 1.0000
N1 0.73616 0.00101 0.74978 1.0000
C1 0.67075 0.04722 0.74986 1.0000
C1 0.62952 0.01582 0.74963 1.0000
C1 0.56495 0.05848 0.74969 1.0000
C1 0.54004 0.13431 0.74998 1.0000
N1 0.47762 0.17740 0.75005 1.0000
C1 0.58222 0.16439 0.75021 1.0000
N1 0.56645 0.23489 0.75053 1.0000
S1 0.63027 0.25054 0.75076 1.0000
N1 0.67787 0.16122 0.75043 1.0000
C1 0.64537 0.12258 0.75015 1.0000
H1 0.64753 0.95796 0.74940 1.0000
H1 0.53494 0.03181 0.74948 1.0000