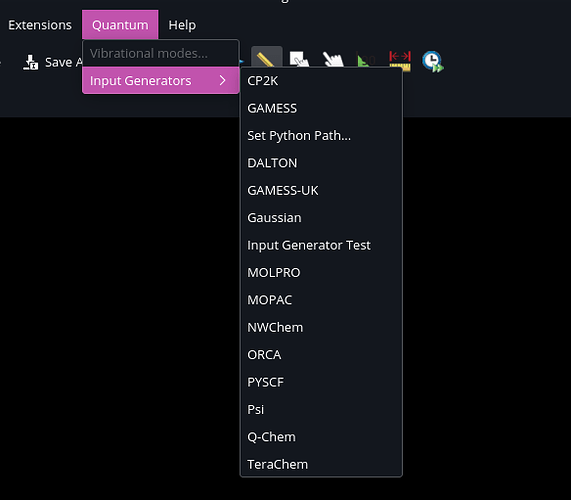
I run Avogadro Flatpak in command line.
flatpak run org.openchemistry.Avogadro2
Which show that it is looking for plugins in these location.
Checking for “inputGenerators” scripts in “/app/share/OpenChemistry/Avogadro/inputGenerators”
Checking for “inputGenerators” scripts in “/usr/share/OpenChemistry/Avogadro/inputGenerators”
Checking for “inputGenerators” scripts in “/usr/share/runtime/share/OpenChemistry/Avogadro/inputGenerators”
Checking for “inputGenerators” scripts in “/run/host/share/OpenChemistry/Avogadro/inputGenerators”
Checking for “inputGenerators” scripts in “/app/bin/…/lib/avogadro2/scripts/inputGenerators”
So I put the inputgenerators plugin here.
/home/.local/share/flatpak/app/org.openchemistry.Avogadro2/current/active/files/share/OpenChemistry/Avogadro/inputGenerators/
which is the same location as “/app/share/OpenChemistry/Avogadro/inputGenerators”
And it found the plugin.
I tried using the plugin downloader. It install the plugins in this location.
Which is not the same place it is looking for. So I recommend you to place the plugins in the directory I show above.
Downloading crystals.zip to /home/.var/app/org.openchemistry.Avogadro2/data/OpenChemistry/Avogadro
Extracting /home/kev/.var/app/org.openchemistry.Avogadro2/data/OpenChemistry/Avogadro/crystals.zip to /home/kev/.var/app/org.openchemistry.Avogadro2/data/OpenChemistry/Avogadro/other/
Extraction successful