@kevinsmia1939
In an installation of Linux Debian 11/bullseye (for less than a week eventually considered stable) and the Xfce4 desktop environment, I followed the instructions and still share the observation by @prasanta13 – there is just a blank, actually just a transparent window:
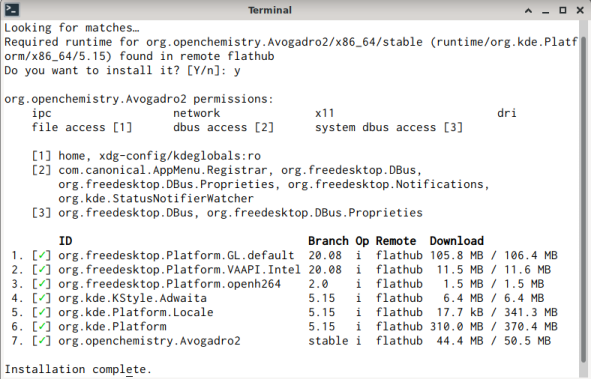
Because I’m not a user of flathub, I perceive the download to get Avogadro comfortable too large. E.g., the download:
with a total of 1.5 GB of space used when eventually installed. So I hope the newer version 1.95 of Avogadro announced here enters soon the package tracker on debichem, e.g. for then (hopefully) functional extensions to prepare jobs for MOPAC, GAMESS, etc.
Till then, it may be better to let apt to manage a local installation of Avogadro (including resolution of any dependencies) than to implant almost a second operating system.