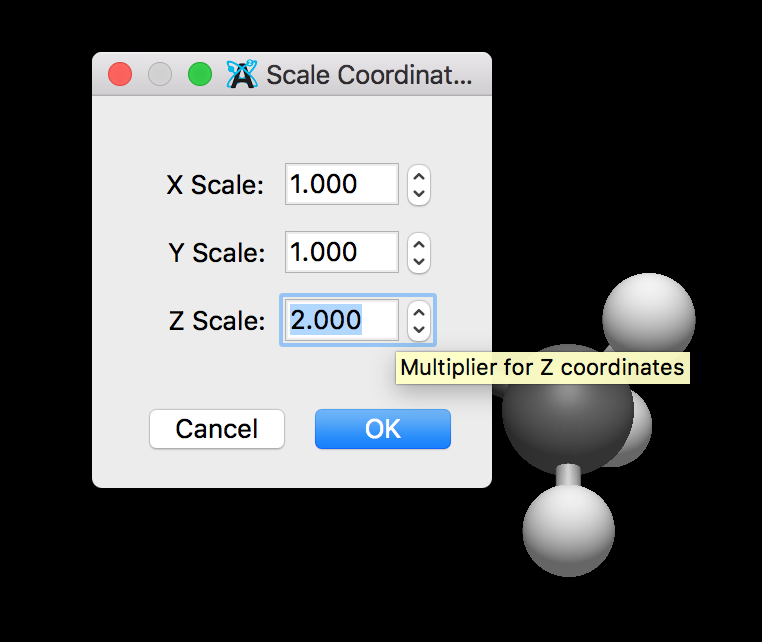
I’m pretty excited that we have an initial draft of (python) workflow scripts. The idea is simple - Avogadro will pass off a molecule to a command-line script, which could be Python or anything else that handles JSON. The script hands back some JSON for a dialog, and when run, it returns a modified molecule to Avogadro.
Undo and redo commands are created automatically.
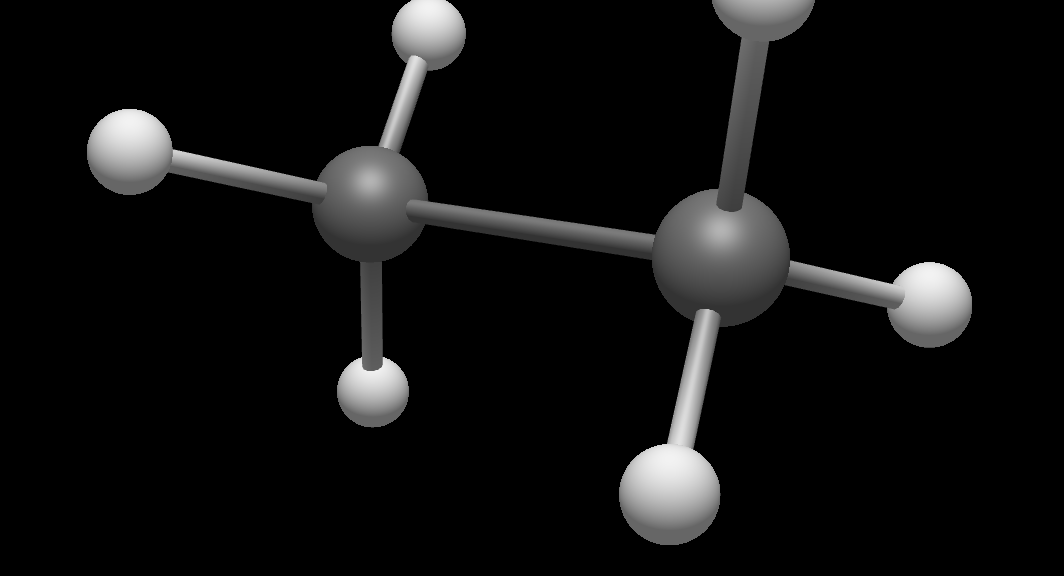
The initial example simply scales the Cartesian coordinates for all atoms. Another example will use a Python package to build nanotubes (https://scikit-nano.org).
I want to ask the community if they have some ideas for use cases. For example, should the scripts be able to return numbers and/or text in addition to the molecule?
- A “flatten molecule” command that makes everything planar, to ensure symmetry
- A “make cluster” command that generates multiple copies of the current molecule along an axis
- An RDKit module that generates conformers
- An ASE module that inserts dopant atoms into octahedral vacancies
Thoughts? We’d like to nail down the API and packaging so multiple scripts can be built up before release.